Author Archive
Appendable saving in R
One of the most crucial problems in HPC is that every error you make have much greater impact than in the normal computing — there is nothing more amusing than finding out that few-day simulation broke few minutes before the end because of an unfortunate value thrown by a random generator, typo in result saving code or just a cluster failure and its result are dissolving abandoned in RAM. Even if all goes OK, often it would be quite nice to have some insight inside the results before the job is completed.
Of course there is a trivial solution to this — frequent saving of preliminary results. Unfortunately this is easier say than done, basically because R’s save cannot append new object to the same file — thus I used to end up either with few giant text files which were somewhere in between something human readable and easily parsable or with a plethora of small RDatas which on the other hand required writing nontrivial script to be reintegrated in something usable (not to mention fuse-ssh freezes and crashes).
To this end I have come up with writing R package intended to expand save with appending; thus rtape was born. How does it work? You simply call
R> rtapeAdd(some_object_worth_saving,"myTape.rtape")
and your first tape appears in the working dir; following rtapeAdds will further populate the file with more objects not removing the previously stored. You can then load the entire stuff with rtapeAsList:
R> rtapeAsList("myTape.rtape")
[[1]]
some_object_worth_saving
[[2]]
some_object_worth_saving2
...
Note that there is no initialization or finalization step — yup, there is no header which makes the tape format simple (for instance you can just cat several files into one, not using R at all) and always ready to use, unfortunately also invalidates any attempt to make it random-readable or mutable in any other way than append / delete all. Though this is not something you expect from a failsafe data dump, do you? Ok, not counting situations when this dump has grown too big to be easily manipulated in the memory; hopefully rtape can also map a function to each tape object storing only current one in the memory, with a help of rtapeLapply and rtape_apply functions. Finally there is also some confidence stuff.
Oh, and rtapeAdd is not (yet) thread-safe — you have been warned (-;
Using OpenMP-ized C code with R
What is OpenMP? Basically a standard compiler extension allowing one to easily distribute calculations over multiple processors in a shared-memory manner (this is especially important when dealing with large data — simple separate-process approach usually requires as many copies of the working data as there are threads, and this may easily be an overkill even in overall size, not to mention the time wasted for copying).
The magic of OpenMP is that once you have a C or Fortran code, in most cases you need nothing more than a few additional compiler flags — thus the code remains as portable and as readable as before the modification. And is usually just nice and simple, not counting few common parallelism traps and some quirks related to the fact we want it to work with R.
In this post I don’t want to make an OMP tutorial (the web is full of them), rather show how to use it with R. Thus, I’ll use a toy example: a function that calculates the cumulative sum in an unnecessary demanding way:
#include <R.h>
#include <Rinternals.h>
SEXP dumbCumsum(SEXP a){
SEXP ans;
PROTECT(a=coerceVector(a,REALSXP));
PROTECT(ans=allocVector(REALSXP,length(a)));
double* Ans=REAL(ans);
double* A=REAL(a);
for(int e=0;e<length(a);e++){
Ans[e]=0.;
for(int ee=0;ee<e+1;ee++)
Ans[e]+=A[ee];
}
UNPROTECT(2);
return(ans);
}
There is only one for loop responsible for most computational time and no race conditions, thus the OMP-ized version will look like this:
#include <R.h>
#include <Rinternals.h>
#include <omp.h>
SEXP dumbCumsum(SEXP a){
SEXP ans;
PROTECT(a=coerceVector(a,REALSXP));
PROTECT(ans=allocVector(REALSXP,length(a)));
double* Ans=REAL(ans);
double* A=REAL(a);
#pragma omp parallel for
for(int e=0;e<length(a);e++){
Ans[e]=0.;
for(int ee=0;ee<e+1;ee++)
Ans[e]+=A[ee];
}
UNPROTECT(2);
return(ans);
}
Time for R-specific improvements; first of all, it is good to give the user an option to select number of cores to use (for instance he has 16 cores and want to use first 8 for one job and next 8 for something else — without such option he would have to stick to sequential execution); yet it is also nice to have some simple option to use the full capabilities of the system. To this end we will give our function an appropriate argument and use OMP functions to comply with it:
#include <R.h>
#include <Rinternals.h>
#include <omp.h>
SEXP dumbCumsum(SEXP a,SEXP reqCores){
//Set the number of threads
PROTECT(reqCores=coerceVector(reqCores,INTSXP));
int useCores=INTEGER(reqCores)[0];
int haveCores=omp_get_num_procs();
if(useCores<=0 || useCores>haveCores) useCores=haveCores;
omp_set_num_threads(useCores);
//Do the job
SEXP ans;
PROTECT(a=coerceVector(a,REALSXP));
PROTECT(ans=allocVector(REALSXP,length(a)));
double* Ans=REAL(ans);
double* A=REAL(a);
#pragma omp parallel for
for(int e=0;e<length(a);e++){
Ans[e]=0.;
for(int ee=0;ee<e+1;ee++)
Ans[e]+=A[ee];
}
UNPROTECT(3);
return(ans);
}
This code will also ensure that the number of threads won’t be larger than the number of physical cores; doing this gives no speedup and comes with a performance loss caused by OS scheduler.
Finally, time to resolve small quirk — R has some code to guard the C call stack from overflows, which is obviously not thread-aware and thus have a tendency to panic and screw the whole R session up when running parallel code. To this end we need to disable it using the trick featured in R-ext. First, we include Rinterface to have an access to the variable with stack limit
#define CSTACK_DEFNS 7 #include "Rinterface.h"
and then set it to almost infinity in the code
R_CStackLimit=(uintptr_t)-1;
Voilà, the stack is now unprotected — the work with R just become a bit more dangerous, but we can run parallel stuff without strange problems. The full code looks like this:
#include <R.h>
#include <Rinternals.h>
#include <omp.h>
#define CSTACK_DEFNS 7
#include "Rinterface.h"
SEXP dumbCumsum(SEXP a,SEXP reqCores){
R_CStackLimit=(uintptr_t)-1;
//Set the number of threads
PROTECT(reqCores=coerceVector(reqCores,INTSXP));
int useCores=INTEGER(reqCores)[0];
int haveCores=omp_get_num_procs();
if(useCores<=0 || useCores>haveCores) useCores=haveCores;
omp_set_num_threads(useCores);
//Do the job
SEXP ans;
PROTECT(a=coerceVector(a,REALSXP));
PROTECT(ans=allocVector(REALSXP,length(a)));
double* Ans=REAL(ans);
double* A=REAL(a);
#pragma omp parallel for
for(int e=0;e<length(a);e++){
Ans[e]=0.;
for(int ee=0;ee<e+1;ee++)
Ans[e]+=A[ee];
}
UNPROTECT(3);
return(ans);
}
Now, time to make sure that R will compile our function with OMP support (and thus make it parallel). To this end, we create a Makevars file (in the src in case of package and in code directory when using dangling object files) with a following contents (for GCC):
PKG_CFLAGS=-fopenmp PKG_LIBS=-lgomp
The first line will trigger parsing OMP pragmas, the latter will link the OMP library with omp_* functions.
We are ready to test our example:
$ R CMD SHLIB omp_sample.c
$ R
> dyn.load('omp_sample.so')
> .Call('dumbCumsum',runif(100000),0L)
Try to run sum system monitor (like htop or GUI one that comes with your desktop environment) and watch your powerful CPU being finally fully utilized (-;
To close with an optimistic aspect, few words about limitations. Don’t even try to run any R code or use features like random number generation or Ralloc inside parallelized blocks — R engine is not thread-safe and thus this will end in a more or less spectacular failure.
Plus of course all issues of parallel programming and OMP itself also apply — but that is a different story.
Challenge alert — material identification
We start yet another series of post — challenge alerts. This series is intended to share news about machine learning or data mining challenges which may be interesting to the members of our community, possibly with some brief introduction to the problem. So if you hear about some contest, notify us on Skewed distribution.
Today about the recent event on TunedIt, where FIND Technologies Inc. asks to develop a method to distinguish various materials based on the passive electromagnetic signals the produce. The supply the participants with 3000 1500-sample time series, each corresponding to a measurement of the electric potential on a surface of one of three materials. Here is a plot of one of given time series:
Half of this set is annotated with a material class and given as a training set, the rest is a test set on which classes must be predicted. This is a `rolling’ challenge, i.e. participants can send many predictions at any time and their results on a preliminary test set (different from a test set used to finally assess their accuracy) are instantly published. Unfortunately, organizers have chosen the preliminary set out of train set samples, so overfitted submissions can get arbitrary high accuracy on the leaderboard. In fact this has happened already, so the real progress remains unknown. After registration, one can download a preliminary report which reveals some technical details about the problem. It also claims that one can obtain circa 70% accuracy in separating each pair of those classes using linear learner on wavelet spectra.
The main downside of the challenge is that is quite frequently regarded as a scam, especially because there is no way of trying to replicate the results from preliminary raport (and the method described therein fails on the challenge data) — more details can be found on the challenge thread on TunedIt forum. Anyway no-one has broke the first, 50% accuracy milestone till now.
The upside is that there are prizes; 1k Canadian $ for breaking 50, 60, 70, 80 and 90% milestone and 40k C$ for braking final goal of 95% accuracy and transferring intelectual rights to FIND.
So, good luck — or have a nice time doing more productive things (=
Two-way CRAN
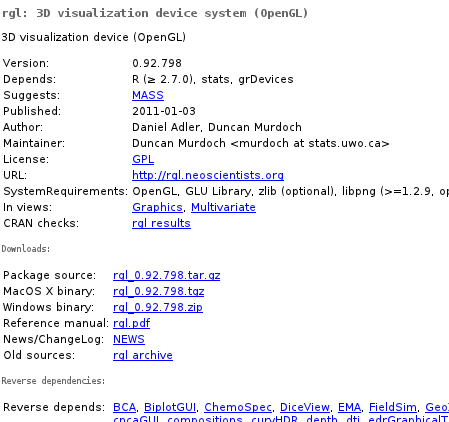
Sooner on later, every useR will manage to exhaust R’s built-in capabilities and land on CRAN looking for his dreamed needle in a haystack of 3k+ contributed packages. Probably most of you already know stuff like Task Views or rseek which make finding something relevant a bit easier than digging the full list or googling, however all methods will eventually lead to a CRAN package page like this:
Ok, but what’s the problem? We have basic info here, sources, manuals, builds, dependencies… Well, let’s compare this to some modern application repositories like Android Market, Mozilla Add-ons or AppStore; we’ll immediately notice lack of any form of user feedback, neither as ratings nor reviews. And such stuff may be quite handy — imagine you have for instance found three packages doing certain thing; surely one of them will be fastest, one least bloated, one most functional and one will best integrate with the rest of your code, but you probably won’t be able to deduce this easily from manuals. Or you’ve found a package from 2003 and you don’t really want to check whether it is an abandoned code dependent on a bunch of obsolete quirks or just a solid code that just didn’t require any modifications. Or you have been using foozifier for years not knowing that bartools’ functionWithConfusingName does the same 50 times faster using 1/1000 of RAM. Or you just thought you can’t baz in R, yet the only problem was that the bazzing package author thought it was called wizzing.
Moreover, this is also useful for package authors — comment is much easier and more reusable way of leaving feedback than e-mail, so you can count on more reports and thus catch more bugs and gather more good feature ideas.
What’s worse with this story is that this is more-less already here; it is called Crantastic and works pretty well, not counting the fact that it would certainly benefit from some more traffic — so, go there and check if you are registered user of all packages you’ve installed and start contributing; it really comes back!
Welcome to the CV blog!
It is almost a year since CrossValidated was launched. Today we start a new activity at CrossValidated — a community blog. It is the fourth (after the main site, meta and chat) place for getting in touch with the community and contributing to it.
To get started, we plan to post series of posts about the following topics:
- Question of the Week Each week there will be a survey on meta in which the users will nominate and elect some recent splendid question — then we’ll ask the author of best answer either to elaborate a bit, write some summary of the whole thread, start a miniseries of posts inspired by the QoW or do something else in this manner.
- Journal Club report There will be some summary of the each JC chat. While the JC has a summer break currently, you can expect some summaries of archived chats.
- R tips&tricks We can not ignore our most popular tag. Hopefully we would manage to attract some people from R community on SO to participate here.
- Challenge alert Announcements (with some introduction) of machine learning or data mining challenges.
Finally the most important thing: remember that this is your place. Anyone can write a post here — just visit Skewed distribution — blog’s chat room and ask for an author account or maybe just suggest some topic or share your critic. We are open to any idea and we hope we’ll manage to make it an useful and avid place for the community.